The challenge
Manuel Fuentes of the Cancer Investigation Centre (CIC), Salamanca was fortunate to collaborate with researchers at Harvard Medical School in developing Nucleic Acid Programmable Protein Arrays (NAPPA). By eliminating the need for recombinant protein production and purification, NAPPA offers a cost-effective and highly reproducible solution for biomarker and drug discovery in tumour and autoimmune pathologies. Having generated results by high-medium density immunoassays in array format, the team were using a Genetix instrument to pin-spot nucleic acids for translation to proteins in situ. The contact-printed arrays lacked reproducibility, displayed cross-contamination and were slow to produce.
The need for speed
Principles behind NAPPA insisted that cDNA and capture antibodies are involved in a master mix which has to be printed within a short time window. Critical success factors meant the current instrument was no longer fit for purpose (Table 1).
| Critical Success Factor | Result with Genetix printer |
|---|---|
| Speed | The short timeframe in which both samples were to be deposited meant that a maximum of 20 slides could be printed in each run |
| Reproducibility | Samples must be printed homogenously to ensure equal protein content. The contact approach did not offer satisfactory reproducibility |
| Sample conservation | A minimum of 10 µg of costly sample was required to print only 20 slides |
| Reliability | Spots must be free of cross contamination |
Table 1: Critical success factors against results with Genetix printer
The Arrayjet solution
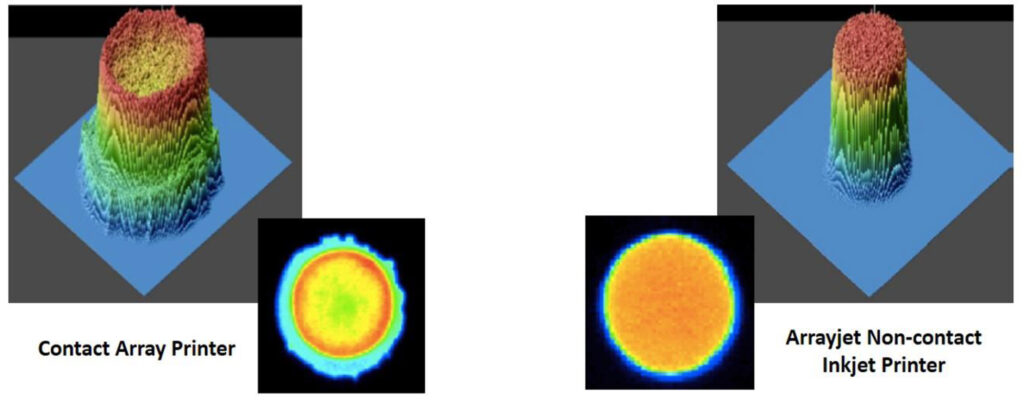
Arrayjet’s non-contact inkjet technology produces spots of noticeably improved morphology compared with pin-spotting systems (Figure 1). As a result, each spot translated to a comparable quantity of protein for reliable data acquisition.
Not only is it more reliable, Arrayjet’s “on-the-fly” method is also the fastest available on the market, enabling deposition of up to 640 features per second. Sample volume requirements are vastly reduced and print runs are largely automated freeing up valuable scientist time.
Transfer to the Arrayjet platform fully satisfied all critical success factors, exceeding customer expectations (Table 2).
| Critical Success Factor | Arrayjet Solution | Benefit |
|---|---|---|
| Speed | “On-the-fly” printing | High-density slides printed without compromising sample |
| Reproducibility | Instruments and components with CV values <5% | Comparable protein quantities translated on each spot |
| Sample conservation | JetSpyder™ liquid handling device | 1.3 µL sample aspiration prints hundreds of slides |
| Reliability | Integrated wash station | Print head and JetSpyder™ do not contribute to contamination |
Summary
Arrayjet’s high-throughput printing can be adapted to suit a variety of applications, including but not limited to:
- Antigen discovery
- Host-pathogen interaction screening
- Biomarker screening
- Epitope mapping
- Antibody validation
- Small molecule library screening
- Hybridoma screening
- Gene expression profiling
Offering both an in-house bioprinting service and a range of five scalable instruments, Arrayjet remain open to any novel screening ideas that test their technology even further.
| Speed | Printing time reduced fourfold leaving scientists free to conduct research |
| Reproducibility | Fivefold increase in slide production |
| Sample conservation | Sample volume demands reduced tenfold |
| Reliability | Significant financial savings associated with automation, printing speed and sample conservation |
“Transfer to Arrayjet technology has had huge benefits for our cancer research and biomarker discovery. Our team can now produce 100 slides in a working day, using just 1 µg of cDNA per gene of interest. The automated process enables us to focus fully on our research knowing arrays will be printed reliably and reproducibly every time.”
Manuel Fuentes, Scientific Researcher at Cancer Investigation Centre


